About Our Research
We develop and deploy cutting-edge systems and synthetic biology approaches to understand the molecular, cellular and ecological principles that shape the structure and function of microbiomes in diverse environments. With these insights, we genetically program microbiomes as next-generation platforms to tackle pressing challenges in personalized medicine, global health, and climate change.
We also have broad interests in understanding the origins of life, the genetic code, metabolism, cancer, and aging, in microbial and mammalian systems.
Ongoing projects include:
Mapping the host and gut microbiome interface in space, across time, and at a strain-level resolution
MaPS-seq and SAMPL-seq are spatial metagenomics methods to study the micron-scale spatial organization of a microbiome (Sheth et al, Nature Biotech 2019) and (Richardson et al, Nature Microbiology 2025). We are applying these methods to identify conserved spatial hubs in the gut microbiome and the metabolic interactions driving such spatial co-association patterns (Urtecho et al, Cell Systems 2024).
DIVERS is a computation method and experimental workflow that quantifies spatial, temporal, and technical variations in microbiome datasets (Ji et al, Nature Methods 2019). We are using this method to understand temporal fluctuations in the gut microbiome.
CAMII is a AI-guided robotic microbiome culturomics system that can analyze and isolate tens of thousands of microbial colonies at a time (Huang et al, Nature Biotech 2023). We are applying this technology to build personalized human microbiome strain collections under healthy and diseased conditions.
Foli-seq is an exfoliome RNA sequencing method to profile the gut transcriptome non-invasively through fecal matter that can assess epithelia, secretory, and immune cells shed from the gut (Huang et al, Nature Biotech 2025). We are using this method to profile the gut transcriptome in IBD, IBS and other gut-related diseases.
Environmental and genetic selection of the gut microbiome
Metabolites in our diets and drugs we use can have a large impact on our gut microbiome (Ricaurte et al, Nature Microbiology 2024). We study how prebiotics and xenobiotics shape the gut microbiome by high-throughput experimentation and deep transcriptomics.
TFUM-seq is a functional metagenomic sequencing method for dissecting genetic factors that enhance microbial fitness and persistence in a microbiome (Yaung et al, Mol Syst Biol 2015). We are using this approach to study now polysaccharide utilization loci’s (PULs) and bile acid metabolism genes serve as key colonization factors.
Synthetic microbial ecosystems can mimic key elements of natural ecology in the lab. We built crossfeeding bacteria to model metabolic interdependency and dynamics (Mee et al, PNAS 2014). Using principles from economics, we developed a model for specialization and mutualistic trade in microbiomes (Tasoff et al, PLoS One 2015; Wall Street Journal 2015).
Microbiome gene therapy to directly program bacterial populations
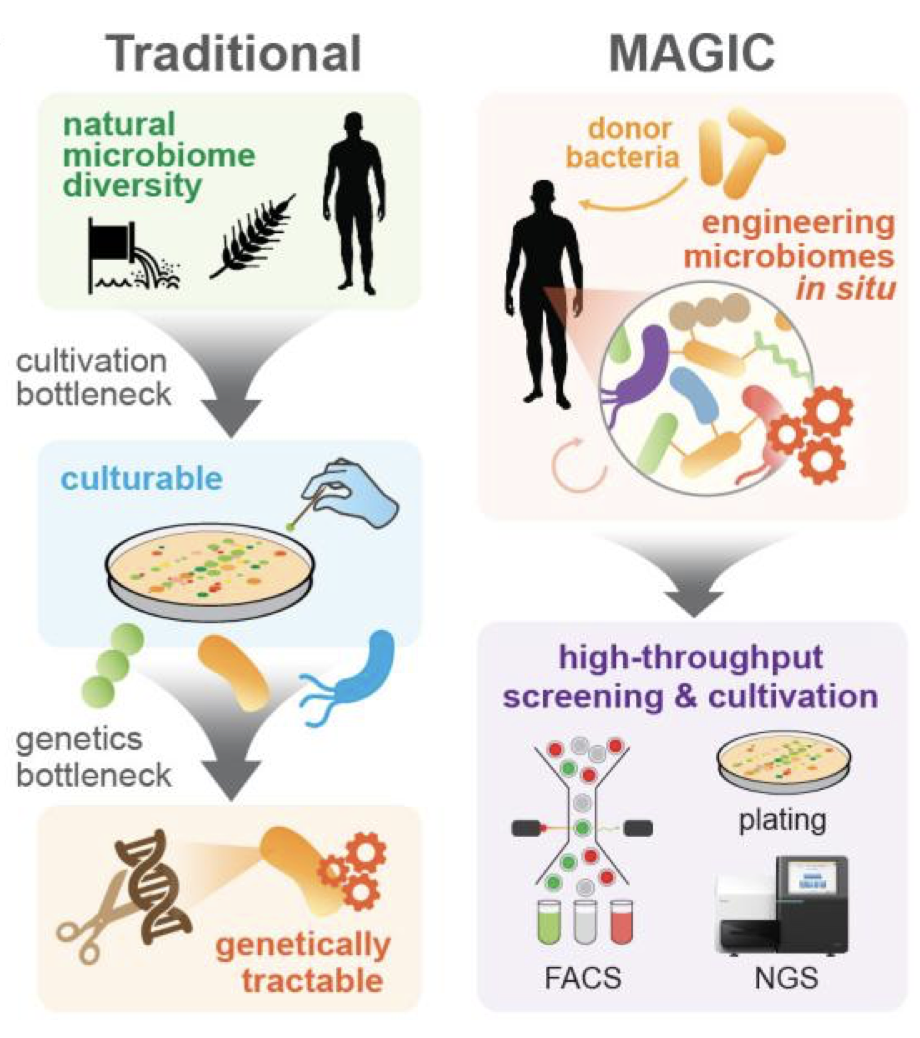
MAGIC is a microbiome engineering method to transfer DNA payloads from a donor cell into recipient bacteria of a microbiome (Ronda et al, Nature Methods 2019). We are using this approach to modify bacteria in situ in the living gut for a variety of therapeutic applications (Sheth et al, Trends in Genetics, 2016).
CRISPR-Cas transposon (CAST) systems enable programmable and precise genomic integration of kilobases of DNA (Vo et al, Nature Biotech 2020). We are using this technology for precise microbiome engineering and editing of pathogens in the gut (Ronda et al, Nature Biomed Eng 2025). We developed Metagenomic Editing (MetaEdit) to engineering commensal microbes in vivo in the gut, including those that cannot be cultured in the lab (Gelsinger et al, Science 2025)
CAMEOS is a computational algorithm to design synthetic sequence entanglements that limit genetic mutations and escape of recombinant DNA (Blazejewski et al, Science 2019). We use this approach to design engineered DNA that have enhanced biocontainment and stability in open environments.
Genetic toolbox of regulatory elements are key for programmable gene expression in microbiomes (Johns et al, Nature Methods 2018). We are characterizing regulatory elements through metagenomic mining and high-throughput reporter assays that enable species-selective and universal genetic circuits (Yim et al, Mol Syst Biol 2019). We further developed CRISPR activator (CRISPRa) systems that are compatible with these regulatory elements (Ho et al, Mol Syst Biol 2020)
Temporal cellular memory in living cells
TRACE is a DNA-based memory system that links a biological signal in the cell or its environment to the generation of new spacers in a CRISPR recording array (Sheth et al, Science 2017). We use this system to build sentinel cells that can track levels of nutrients, toxins, and pathogens in the environment (Sheth et al, Nature Reviews Genetics 2018).
DRIVES is the first direct digital-to-biological data storage system that takes binary electrical stimuli and converts them to genomically recorded CRISPR spacers (Yim et al, Nature Chemical Biology 2021). We are using this approach to build cellular memory systems that can interface with computers directly.
CRISPR-spacer recording allows active tracking of gene flow of mobile DNA in a microbiome (Munck, Nature Communications 2020). We are using this method to study the mobilome of the gut and soil environments.
Engineering microbes for biomaterial, biorefinement, and biocontrol applications
Fungal mycelium and bacteria can grow on lignocellulosic feedstocks to form moldable, foldable, and regenerative living structures (McBee et al, Nature Materials 2021). We use bioprospecting and synthetic biology to engineer these living biocomposites with new biosynthetic and sensing-reporting capabilities.
Microbial biofilms can resist colonization of pathogens. We are developing microbial biofilms that can repel the growth of human-associated pathogens including black molds.
Extremophiles have unique abilities to concentrate and process metabolites and minerals. We are isolating and engineering microbes that can refine high-value minerals from the environment.
Gene synthesis, Genome Project-write (GP-write), Genetic code editing
MEGAA is a gene synthesis method that enables cheap and rapid construction of genetic variants (Liu et al, Nature Methods 2023). This technology is being used to generate SARS-CoV2 spike protein variants for pandemic surveillance (Liu et al, Nature 2021; Iketani et al, Nature 2022; Wang et al, Cell 2022) and viral variants for gene delivery applications.
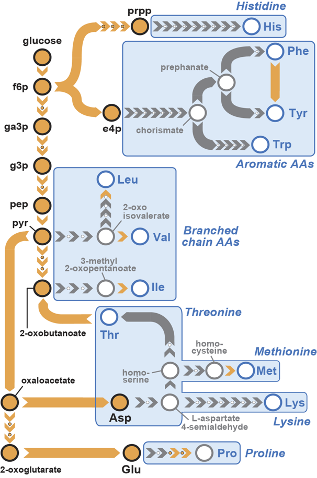
Pathways for biosynthesis of essential amino acids have been lost from the Animalia kingdom for over 500 million years. Using synthetic genomics strategies and as a part of the GP-write effort (Boeke et al, Science 2016), we are resurrecting these pathways in mammalian cells to augment their metabolism. We described the successful resurrection of a bacterial valine biosynthesis pathway in a mammalian cell line (Trolle et al, eLife 2022).
MAGE (Wang et al, Nature 2009) and CAGE (Isaacs et al, Science, 2011) are genome editing and assembly technologies that helped produce genomically recoded organisms (Lajoie et al, Science 2013). Using these and additional approaches (Wang et al, Nature Methods, 2012; Kelsic et al, Cell Systems, 2016) along with computational protein design, we are exploring genetic code designs that push the limits of biology.
We use a variety of model organisms and systems including:
Bacteria: aerobic and anaerobic gut bacteria, soil microbes and extremophiles
Eukarya: yeast, fungi, mammalian cells
Animal models: conventional and gnotobiotic mice
Clinical specimens
Funding
We are very grateful for the generous support of our research by the following agencies and foundations: NIH-OD, NIGMS, NIAID, NHGRI, NIDDK, NCI, NIBIB, ONR, ARO, AFRO, DARPA, NSF, DOE, Burroughs Wellcome Fund, Sloan Foundation, Bill & Melinda Gates Foundation, Crohn’s and Colitis Foundation, Schaefer Scholars Program, Hirschl Trust, Takeda Research, CZ Biohub NY.