* denote shared co-authorship, # denote co-corresponding/co-senior authorship.
Publications
Selected Recent Highlights
Fecal exfoliome sequencing captures immune dynamics of the healthy and inflamed gut
Huang Y*, Sun Y*, Ronda C, Mavros CF, Li J, Jacobse J, Huang LH, Resnick SJ, Giddins M, Freedberg DE, Chavez A, Goettel JA, Wang HH
Nature Biotechnology doi: 10.1038/s41587-025-02894-4 (2025) [pdf]
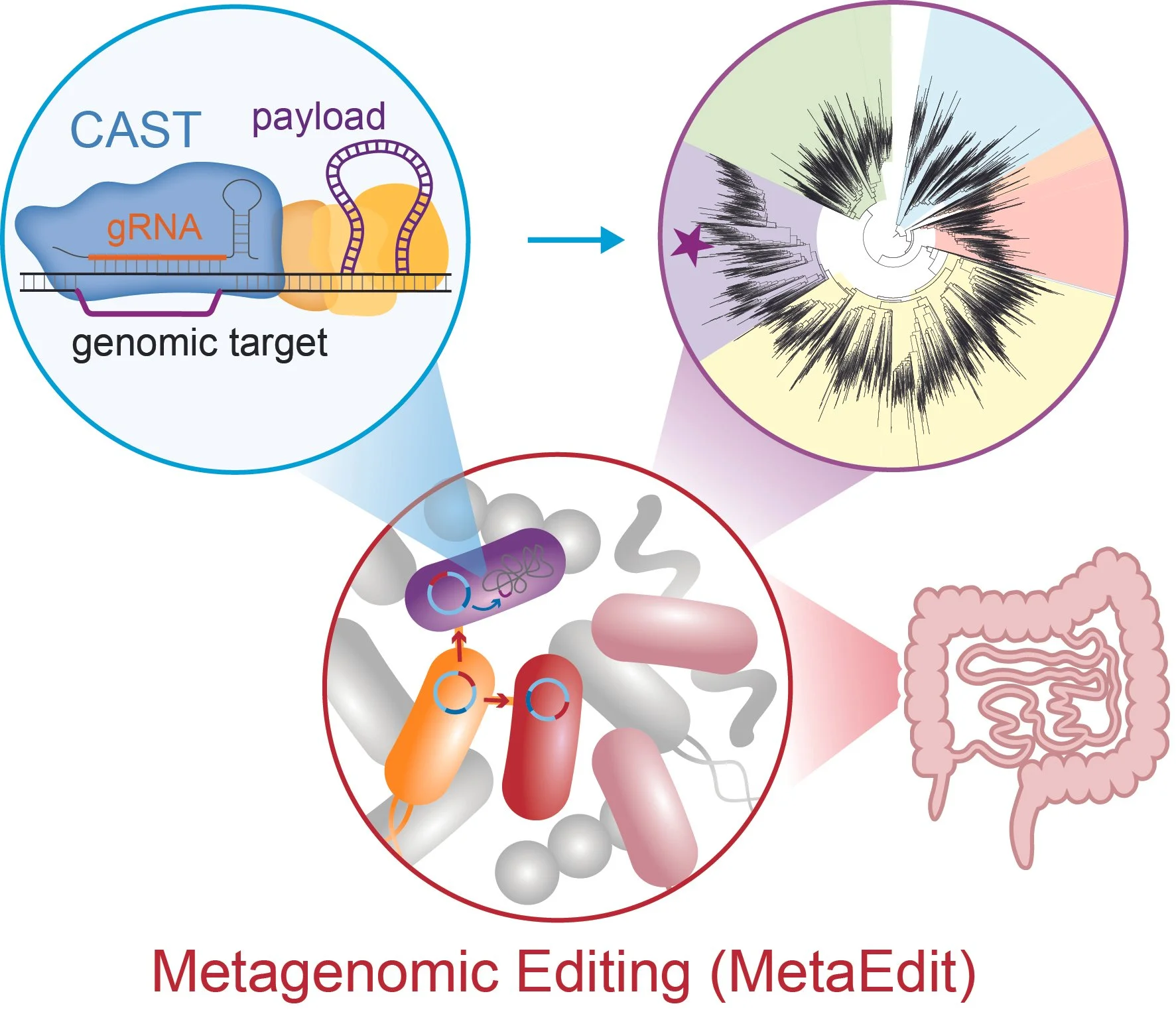
Metagenomic editing of commensal bacteria in vivo using CRISPR-associated transposases.
Gelsinger D, Ronda C, Ma J, Kar OB, Edwards M, Huang Y, Mavros C, Sun Y, Perdue T, Vo PL, Ivanov II#, Sternberg SH#, Wang HH#
Science doi: 10.1126/science.adx7604 (2025) [pdf]. Also see Perspective piece in Science [here]
Engineered probiotic restores GLP-1 signaling to ameliorate fiber-deficiency exacerbated colitis
Brockmann L*, Ronda C*, Schwanz LT, Qu Y, Shneider DW, Mavros CF, Ivanov II, Bhagat G, Wang HH
Science Advances doi: 10.1126/sciadv.adx6869 (2025) [pdf]
Precise virulence inactivation using a CRISPR-associated transposase for combating Enterobacteriaceae gut pathogens
Ronda C*, Perdue T*, Schwanz L, Gelsinger DR, Brockmann L, Kaufman A, Huang Y, Sternberg SH, Wang HH
Nature Biomedical Engineering doi: 10.1038/s41551-025-01453-1 (2025) [pdf][SI]
SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome
Richardson M, Zhao S, Lin L, Sheth RU, Qu Y, Lee J, Moody T, Ricaurte D, Huang Y, Velez-Cortes F, Urtecho G, Wang HH
Nature Microbiology doi: 10.1038/s41564-024-01914-4 (2025) [pdf]
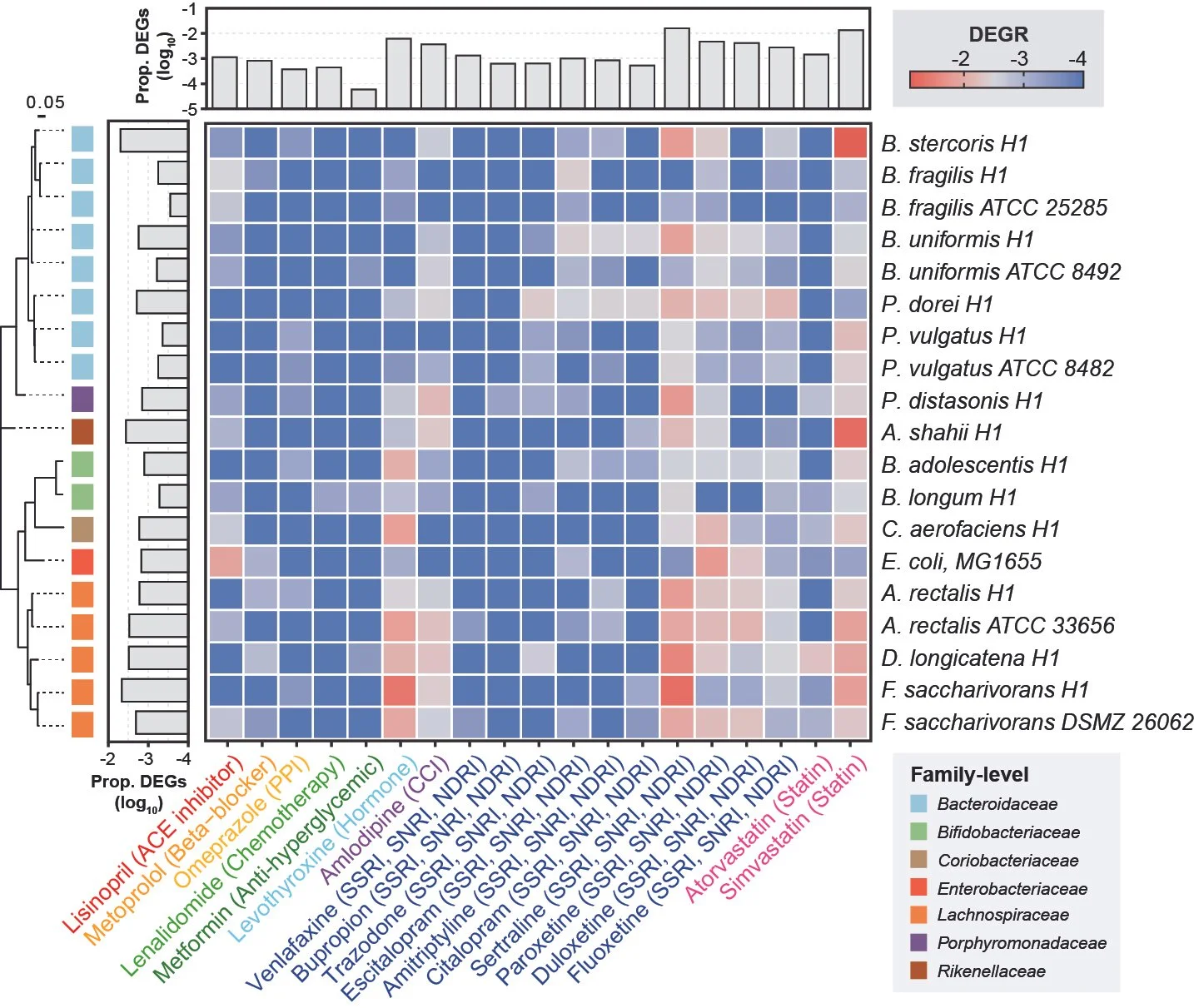
High-throughput transcriptomics of 409 bacteria–drug pairs reveals drivers of gut microbiota perturbation
Ricaurte D*, Huang Y*, Sheth RU, Gelsinger DR, Kaufman A, Wang HH
Nature Microbiology 9:561-575 (2024) [pdf]
Fast and efficient template-mediated synthesis of genetic variants
Liu L*, Huang Y*, Wang HH
Nature Methods 20:841-848 (2023) [pdf][protocol]
High-throughput microbial culturomics using automation and machine learning
Huang Y*, Sheth RU*, Zhao S, Cohen L, Dabaghi K, Moody T, Sun Y, Ricaurte D, Richardson M, Velez-Cortes F, Blazejewski T, Kaufman A, Ronda C, Wang HH
Nature Biotechnology 41:1424-1433 (2023) [pdf]
Resurrecting essential amino acid biosynthesis in a mammalian cell
Trolle J*, McBee RM*, Kaufman A, Pinglay S, Berger H, German S, Liu L, Shen MJ, Guo X, Martin JA, Pacold M, Jones DR, Boeke JD#, Wang HH#
eLife doi: 10.7554/eLife.72847 (2022) [pdf]
Engineering living and regenerative fungal–bacterial biocomposite structures
McBee RM, Lucht M, Mukhitov N, Richardson M, Srinivasan T, Meng D, Chen H, Kaufman A, Reitman M, Munck C, Schaak D#, Voigt C, Wang HH#
Nature Materials 21:471-478 (2022) [pdf]
Robust direct digital-to-biological data storage in living cells
Yim SS, McBee RM, Song AM, Huang Y, Sheth RU, Wang HH
Nature Chemical Biology 17:246-253 (2021) [pdf]
Synthetic sequence entanglement augments stability and containment of genetic information in cells
Blazejewski T*, Ho HI*, Wang HH.
Science 365:595-598 (2019) [pdf]
Spatial metagenomic characterization of microbial biogeography in the gut
Sheth RU, Li M, Jiang W, Sims PA, Leong KW, Wang HH.
Nature Biotechnology 37:877-883 (2019) [pdf]
Metagenomic engineering of the mammalian gut microbiome in situ
Ronda C*, Chen SP*, Cabral V*, Yaung SJ, Wang HH
Nature Methods 16:167-170 (2019) [pdf]
Metagenomic mining of regulatory elements enables programmable species-selective gene expression
Johns NI*, Gomes ALC*, Yim SS, Yang A, Blazejewski T, Smillie CS, Smith MB, Alm EJ, Kosuri S, Wang HH
Nature Methods 15:323-329 (2018) [pdf]
Multiplex recording of cellular events over time on CRISPR biological tape
Sheth RU, Yim SS, Wu FL, Wang HH
Science 358:1457-1461 (2017) [pdf]
Published (Full List)
Fecal exfoliome sequencing captures immune dynamics of the healthy and inflamed gut
Huang Y*, Sun Y*, Ronda C, Mavros CF, Li J, Jacobse J, Huang LH, Resnick SJ, Giddins M, Freedberg DE, Chavez A, Goettel JA, Wang HH
Nature Biotechnology doi: 10.1038/s41587-025-02894-4 (2025) [pdf]
Metagenomic editing of commensal bacteria in vivo using CRISPR-associated transposases.
Gelsinger D, Ronda C, Ma J, Kar OB, Edwards M, Huang Y, Mavros C, Sun Y, Perdue T, Vo PL, Ivanov II#, Sternberg SH#, Wang HH#
Science doi: 10.1126/science.adx7604 (2025) [pdf]. Also see perspective piece in Science [here]
Engineered probiotic restores GLP-1 signaling to ameliorate fiber-deficiency exacerbated colitis
Brockmann L*, Ronda C*, Schwanz LT, Qu Y, Shneider DW, Mavros CF, Ivanov II, Bhagat G, Wang HH
Science Advances doi: 10.1126/sciadv.adx6869 (2025) [pdf]
Precise virulence inactivation using a CRISPR-associated transposase for combating Enterobacteriaceae gut pathogens
Ronda C*, Perdue T*, Schwanz L, Gelsinger DR, Brockmann L, Kaufman A, Huang Y, Sternberg SH, Wang HH
Nature Biomedical Engineering doi: 10.1038/s41551-025-01453-1 (2025) [pdf][SI]
Loss of FXR or Bile Acid-dependent Inhibition Accelerate Carcinogenesis of Gastroesophageal Adenocarcinoma
Baumeister T, Proaño-Vasco A, Metwaly A, Kleigrewe K, Kuznetsov A, Schömig LR, Borgmann M, Khiat M, Anand A, Strangmann J, Böttcher K, Haller D, Dunkel A, Somoza V, Reiter S, Meng C, Thimme R, Schmid RM, Patil DT, Burgermeister E, Quante M
Cellular and Molecular Gastroenterology and Hepatology 19(8):101505 (2025) [pdf]
SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome
Richardson M, Zhao S, Lin L, Sheth RU, Qu Y, Lee J, Moody T, Ricaurte D, Huang Y, Velez-Cortes F, Urtecho G, Wang HH
Nature Microbiology doi: 10.1038/s41564-024-01914-4 (2025) [pdf]
Diary of a cell in DNA ‘chyrons’
Chen C, Wang HH.
Nature Chemical Biology doi: 10.1038/s41589-024-01814-y (2025) [pdf]
Bacteria engineered to produce serotonin modulate host intestinal physiology
Mavros CF, Bongers M, Neergaard FBF, Cusimano F, Sun Y, Kaufman A, Richardson M, Kammler S, Kristensen M, Sommer MOA, Wang HH.
ACS Synth Biol 13:4002−4014 (2024) [pdf]
Spatiotemporal dynamics during niche remodeling by super-colonizing microbiota in the mammalian gut
Urtecho G, Moody T,∙Huang Y, Sheth RU, Richardson M, Descamps HC, Kaufman A, Lekan O, Zhang Z, Velez-Cortes F, Qu Y, Cohen L, Ricaurte D, Gibson TE, Gerber GK, Thaiss CA, Wang HH.
Cell Systems 15:1-16 (2024) [pdf]
Context-dependent role of group 3 innate lymphoid cells in mucosal protection
Araujo LP, Edwards M, Irie K, Huang Y, Kawano Y, Tran A, De Michele S, Bhagat G, Wang HH, Ivanov II.
Science Immunology 9(98):eade7530 (2024) [pdf]
GENTANGLE: integrated computational design of gene entanglements
Martí JM, Hsu C, Rochereau C, Xu C, Blazejewski T, Nisonoff H, Leonard SP, Kang-Yun CS, Chlebek J, Ricci DP, Park D, Wang HH, Listgarten J, Jiao Y, Allen JE.
Bioinformatics 40(7):btae380 (2024) [pdf]
High-throughput transcriptomics of 409 bacteria–drug pairs reveals drivers of gut microbiota perturbation
Ricaurte D*, Huang Y*, Sheth RU, Gelsinger DR, Kaufman A, Wang HH
Nature Microbiology 9:561-575 (2024) [pdf]
Bacterial genome engineering using CRISPR-associated transposases
Gelsinger DR, Vo PH, Klompe SE, Ronda C, Wang HH, Sternberg SH
Nature Protocols doi: 10.1038/s41596-023-00927-3 (2024) [pdf]
Intestinal microbiota-specific Th17 cells possess regulatory properties and suppress effector T cells via c-MAF and IL-10
Brockmann L, Tran A, Huang Y, Edwards M, Ronda C, Wang HH, Ivanov II
Immunity doi: 10.1016/j.immuni.2023.11.003 (2023) [pdf]
Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike
Wang Q, Guo Y, Liu L, Schwanz LT, Li Z, Nair MS, Ho J, Zhang RM, Iketani S, Yu J, Huang Y, Qu Y, Valdez Y, Lauring AS, Huang Y, Gordon A, Wang HH, Liu L, Ho DD
Nature doi: 10.1038/s41586-023-06750-w (2023) [pdf]
Fast and efficient template-mediated synthesis of genetic variants
Liu L*, Huang Y*, Wang HH
Nature Methods 20:841-848 (2023) [pdf][protocol]
High-throughput microbial culturomics using automation and machine learning
Huang Y*, Sheth RU*, Zhao S, Cohen L, Dabaghi K, Moody T, Sun Y, Ricaurte D, Richardson M, Velez-Cortes F, Blazejewski T, Kaufman A, Ronda C, Wang HH
Nature Biotechnology 41:1424-1433 (2023) [pdf]
Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants
Wang Q, Iketan S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, Valdez R, Lauring AS, Sheng Z, Wang HH, Gordon A, Liu L, Ho DD
Cell doi: 10.1016/j.cell.2022.12.018 (2022) [pdf]
Characterization and spatial mapping of the human gut metasecretome
Velez-Cortes F, Wang HH
mSystems e0071722. doi: 10.1128/msystems.00717-22 (2022) [pdf]
Resurrecting essential amino acid biosynthesis in a mammalian cell
Trolle J*, McBee RM*, Kaufman A, Pinglay S, Berger H, German S, Liu L, Shen MJ, Guo X, Martin JA, Pacold M, Jones DR, Boeke JD#, Wang HH#
eLife doi: 10.7554/eLife.72847 (2022) [pdf]
Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome
Kawano Y, Edwards E*. Huang Y*, Bilate AM, Araujo LP, Tanoue T, Atarashi K, Ladinsky MS, Reiner SL, Wang HH, Mucida D, Honda K, Ivanov II
Cell doi: 10.1016/j.cell.2022.08.005 (2022) [pdf]
Decreased gut microbiome tryptophan metabolism and serotonergic signaling in patients with persistent mental health and gastrointestinal symptoms after COVID-19
Blackett JW, Sun Y, Purpura L, Margolis KG, Elkind MSV, O'Byrne S, Wainberg M, Abrams JA, Wang HH, Chang L, Freedberg DE
Clin Transl Gastroenterol doi: 10.14309/ctg.0000000000000524 (2022) [pdf]
Antibody evasion properties of SARS-CoV-2 Omicron sublineages
Iketani S*, Liu L*, Guo Y*, Liu L*, Chan JFW, Huang Y, Wang M, Luo Y, Yu J, Chu H, Chik KKH, Yuen TTT, Yin MT, Sobieszczyk ME, Huang Y, Yuen KY, Wang HH, Sheng Z, Ho DD.
Nature doi: 10.1038/s41586-022-04594-4 (2022) [pdf]
Engineering temporal dynamics in microbial communities
Ronda C, Wang HH.
Current Opinion in Microbiology 65:47-55 (2022) [pdf]
Cryo-EM structure of the SARS-CoV-2 Omicron spike
Cerutti G*, Guo Y*, Liu L*, Liu L*, Zhang Z, Luo Y, Huang Y, Wang H.H., Ho DD, Sheng Z, Shapiro L.
Cell Reports doi: 10.1016/j.celrep.2022.110428 (2022) [pdf]
Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2
Liu L*, Iketani S*, Guo Y*, Chan JFW*, Wang M*, Liu L*, Luo Y, Chu H, Huang Y, Nair MS, Yu J, Chik KKJ, Yuen TTT, Yoon C, To KKW, Chen H, Yin MT, Sobieszczyk ME, Huang X, Wang HH, Sheng Z, Yuen KY, Ho DD
Nature 602:676-681 (2022) [pdf]
Engineering living and regenerative fungal–bacterial biocomposite structures
McBee RM, Lucht M, Mukhitov N, Richardson M, Srinivasan T, Meng D, Chen H, Kaufman A, Reitman M, Munck C, Schaak D#, Voigt C, Wang HH#
Nature Materials 21:471-478 (2022) [pdf]
Extensive regulation of enzyme activity by phosphorylation in Escherichia coli
Schastnaya E, Nakic ZR, Gruber C, Doubleday P, Krishnan A, Johns NI, Park J, Wang HH, Sauer U
Nature Communications 12: 5650 (2021) [pdf]
Systematic dissection of σ70 sequence diversity and function in bacteria
Park J, Yim SS, Wang HH
Cell Reports 36(8):109590 (2021) [pdf]
High-throughput transcriptional characterization of regulatory sequences from bacterial biosynthetic gene clusters
Park J, Yim SS, Wang HH
ACS Synthetic Biology 10(8):1859-1873 (2021) [pdf]
Fusobacterium nucleatum secretes amyloid-likeFadA to enhance pathogenicity
Meng Q, Gao Q, Mehrazarin S, Tangwanichgapong K, Wang Y, Huang Y, Pan Y, Robinson S, Liu S, Zangiabadi A, Lux R, Papapanou PN, Guo XE, Wang HH, Berchowitz LE, Han YW.
EMBO Rep 22:e52891 (2021) [pdf]
Exploiting interbacterial antagonism for microbiome engineering
Yim SS, Wang HH
Current Opinion in Biomedical Engineering 19:100307 (2021) [pdf]
Robust direct digital-to-biological data storage in living cells
Yim SS, McBee RM, Song AM, Huang Y, Sheth RU, Wang HH
Nature Chemical Biology 17:246-253 (2021) [pdf]
CRISPR RNA-guided integrases for high-efficiency and multiplexed bacterial genome engineering
Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH, Sternberg SH
Nature Biotechnology 39:480-489 (2020) [pdf]
Protecting Linear DNA Templates in Cell-Free Expression Systems from Diverse Bacteria
Yim SS, Johns NI, Noireaux V, Wang HH
ACS Synthetic Biology 9(10):2851-2855 (2020) [pdf]
Programmable and portable CRISPR-Cas transcriptional activation in bacteria
Ho HI, Fang JR, Cheung J, Wang HH
Molecular Systems Biology 16:e9427 (2020) [pdf]
The effect of short-course antibiotics on the resistance profile of colonizing gut bacteria in the ICU: a prospective cohort study
Munck C, Sheth RU, Cuaresma E, Weidler J, Stump SL, Zachariah P, Chong DH, Uhlemann AC, Abrams JA, Wang HH, Freedberg DE
Critical Care 24(1):404 (2020) [pdf]
Genome and sequence determinants governing the expression of horizontally acquired DNA in bacteria
Gomes ALC*, Johns NI*, Yang A, Velez-Cortes F, Smillie CS, Smith MB, Alm EJ, Wang HH
The ISME Journal 14:2347-2357 (2020) [pdf]
Impact of fiber-based enteral nutrition on the gut microbiome of ICU patients receiving broad-spectrum antibiotics: a randomized pilot trial
Freedberg DE, Messina M, Lynch E, Tess M, Miracle E, Chong DH, Wahab R, Abrams JA, Wang HH, Munck C.
Critical Care Explorations 2(6):e0135 (2020) [pdf]
Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform
Munck C*, Sheth RU*, Freedberg DE, Wang HH
Nature Communications 11:95 (2020) [pdf]
Scalable and cost-effective ribonuclease-based rRNA depletion for bacterial transcriptomics
Huang Y*, Sheth RU*, Kaufman AM, Wang HH
Nucleic Acids Research 48(4):e20 (2019) [pdf]
An engineered Cas-Transposon system for programmable and site-directed DNA transpositions
Chen SP, Wang HH
The CRISPR Journal doi: 10.1089/crispr.2019.0030 (2019) [pdf]
Molecular function limits divergent protein evolution on planetary timescales
Konate M, Plata G, Park J, Usmanova DR, Wang HH, Vitkup D
Elife 8:e39705 (2019) [pdf]
Multiplex transcriptional characterizations across diverse bacterial species using cell‐free systems
Yim SS*, Johns NI*, Park J, Gomes ALC, McBee RM, Richardson M, Ronda C, Chen SP, Garenne D, Noireaux V, Wang HH
Molecular Systems Biology 15:e8875 (2019) [pdf]
Synthetic sequence entanglement augments stability and containment of genetic information in cells
Blazejewski T*, Ho HI*, Wang HH.
Science 365:595-598 (2019) [pdf]
Spatial metagenomic characterization of microbial biogeography in the gut
Sheth RU, Li M, Jiang W, Sims PA, Leong KW, Wang HH.
Nature Biotechnology 37:877-883 (2019) [pdf]
Quantifying spatiotemporal variability and noise in absolute microbiota abundances using replicate sampling
Ji BW*, Sheth RU*, Dixit PD, Huang Y, Kaufman A, Wang HH#, Dennis Vitkup#
Nature Methods 16:731-736 (2019) [pdf]
Metagenomic engineering of the mammalian gut microbiome in situ
Ronda C*, Chen SP*, Cabral V*, Yaung SJ, Wang HH
Nature Methods 16:167-170 (2019) [pdf]
Characterizing posttranslational modifications in prokaryotic metabolism using a multiscale workflow
Brunk E, Chang RL, Xia J, Hefzi H, Yurkovich JT, Kim D, Buckmiller E , Wang HH, Cho BK , Yang C , Palsson BO, Church GM, Lewis NE
Proc Natl Acad Sci USA 115(43):11096-11101 (2018) [pdf]
DNA-based memory devices for recording cellular events
Sheth R, Wang HH
Nature Reviews Genetics 19:718–732 (2018) [pdf]
Metagenomic mining of regulatory elements enables programmable species-selective gene expression
Johns NI*, Gomes ALC*, Yim SS, Yang A, Blazejewski T, Smillie CS, Smith MB, Alm EJ, Kosuri S, Wang HH
Nature Methods 15:323-329 (2018) [pdf]
Systematic and synthetic approaches to rewire regulatory networks
Park J, Wang HH
Curr Opin Syst Biol 8:90-96 (2018) [pdf]
Multiplex recording of cellular events over time on CRISPR biological tape
Sheth RU, Yim SS, Wu FL, Wang HH
Science 358:1457-1461 (2017) [pdf]
RNA structural determinants of optimal codons revealed by MAGE-Seq
Kelsic ED*, Chung H*, Cohen N, Park J, Wang HH#, Kishony R#
Cell Systems 3(6):563-571 (2016) [pdf]
A high-throughput strategy for dissecting mammalian genetic interactions
Stockman VB, Ghamsari L, Lasso G, Honig B, Shapira SD, Wang HH
PLoS One 11(12):e0167617 (2016) [pdf]
The genome project-write
Boeke JD, Church GM, Hessel A, Kelly NJ, et al.
Science 353:126-127 (2016) [pdf]
The role of genome accessibility in transcription factor binding in bacteria
Gomes ALC, Wang HH
PLoS Comput Biol 12(4):e1004891 (2016). [pdf]
Global rebalancing of cellular resources by pleiotropic point mutations illustrates a multi-scale mechanism of adaptive evolution
Utrilla J, O'Brien EJ, Chen K, McCloskey D, Cheung J, Wang HH, Armenta-Medina D, Feist AM, Palsson BO
Cell Systems 2:260-271 (2016) [pdf]
Principles for designing synthetic microbial communities
Johns NI, Blazejewski T, Gomes ALC, Wang HH
Curr Opin Microbiol 31:146-153 (2016) [pdf]
Manipulating bacterial communities by in situ microbiome engineering
Sheth RU, Cabral V, Chen SP, Wang HH
Trends in Genetics 32:189-200 (2016) [pdf]
Challenges in microbial ecology: building predictive understanding of community function and dynamics
Widder S., Allen RJ , Pfeiffer T, Curtis TP, et al.
The ISME Journal 10(11):2557-2568 (2016) [pdf]
Proton pump inhibitors alter specific taxa in the human fecal microbiome: results of a crossover trial
Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Susan Whittier S, Wang TC, Wang HH#, Abrams JA#
Gastroenterology 149:883-885 (2015) [pdf]
An economic framework of microbial trade
Tasoff J, Mee MT, Wang HH
PLoS One 10(7):e0132907 (2015) [pdf]
Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics
Yaung SJ, Deng L, Li N, Braff JL, Church GM, Bry L, Wang HH#, Gerber GK#
Molecular Systems Biology 11:788 (2015) [pdf] Also see MSB news & views commentary [here].
Prediction of resistance development against drug combinations by collateral responses to component drugs
Munck C, Gumpert HK, Wallin AIN, Wang HH, Sommer MOA
Science Translational Medicine 262(6):262ra156 (2014) [pdf]
Transfer of noncoding DNA drives regulatory rewiring in bacteria
Orena Y, Smith MB, Johns NI, Zeevia MK, Birand D, Rond EZ, Coranderf J, Wang HH, Alm EJ, Pupko T
Proc Natl Acad Sci USA 111(45): 16112-16117 (2014) [pdf]
Syntrophic exchange in synthetic microbial communities
Mee MT, Collins JJ, Church GM#, Wang HH#
Proc Natl Acad Sci USA 111:E2149-E2156 (2014) [pdf]
MODEST: a web-based design tool for oligonucleotide-mediated genome engineering and recombineering
Bonde MT, Klausen MS, Anderson MV, Wallin AIN, Wang HH#, Sommer MOA#
Nucleic Acids Research 42(W1):W408-W415 (2014) [pdf]
Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides
Bonde MT*, Kosuri S*, Genee HJ, Sarup-Lytzen K, Church GM#, Sommer MOA#, Wang HH#
ACS Synthetic Biology doi: 10.1021/sb5001565 (2014) [pdf]
Recent progress in engineering human-associated microbiomes
Yaung S, Church G, Wang HH
Method Mol Bio 1151:3-25 (2014) [pdf]
Yeast Oligo-mediated Genome Engineering (YOGE)
DiCarlo JE, Conley AJ, Penttilä M, Jäntti J, Wang HH#, Church GM#
ACS Synthetic Biology 342: 357-360 (2013) [pdf]
Genomically recoded organisms expand biological functions
Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA,Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ
Science 342: 357-360 (2013) [pdf]
Genome-scale engineering for systems and synthetic Biology
Esvelt KM#, Wang HH#
Molecular Systems Biology 9:641 (2013) [pdf]
Applications of engineered synthetic ecosystems
Wang HH, Mee MT, Church GM
Synthetic Biology: Tools and Applications, Editor: Huimin Zhao, Elsevier, (2013) [pdf]
Pre-2013 Publications
Engineering ecosystems and synthetic ecologies
Mee M, Wang HH
Mol BioSys 8:2470-2483, (2012) [pdf]
Genome-scale promoter engineering by co-selection MAGE
Wang HH*, Kim HB*, Cong L, Jeong JH, Bang D, Church GM
Nature Methods 9: 591-593 (2012) [pdf]
Multipliexed in vivo tagging of enzyme ensembles with MAGE for in vitro single-pot multi-enzyme catalysis
Wang HH*, Huang P*, Xu G, Marbelstone A, Li J, Forster T, Jewett MC, Church GM.
ACS Synth Biol 1:43-52 (2012) [pdf]
Enhanced multiplex genome engineering through cooperative oligonucleotide co-selection
Carr PA*, Wang HH*, Sterling B*, Isaacs FJ, Xu G, Kraal L, Bang D, Jacobson J, Church GM
Nucleic Acids Res doi: 10.1093/nar/gks455 (2012) [pdf]
Improving lambda red genome engineering via rational removal of endogenous nucleases
Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM. Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM
PLoS One 7(9): e44638. doi:10.1371/journal.pone.0044638 (2012) [pdf]
Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering
Wang HH, Church GM
Methods Enzymol 498:409-26 (2011) [pdf]
Precise manipulation of chromosomes in vivo enables genome-wide codon replacement
Isaacs FJ*, Carr PA*, Wang HH*, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM
Science 333: 348-53 (2011) [pdf]
Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion
Wang HH, Xu G, Vonner AJ, Church G
Nucleic Acids Res 39(16): 7336-47 (2011) [pdf]
Synthetic Genomes for Synthetic Biology
Wang HH
J Mol Cell Biol 2(4): 178-179 (2010) [pdf]
Programming cells by multiplex genome engineering and accelerated evolution
Wang HH*, Isaacs FJ*, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM
Nature 460:894-8 (2009) [pdf]
Physiological noise in MR images: an indicator of the tissue response to ischemia?
Wang HH, Menezes NM, Zhu MW, Ay H, Koroshetz WJ, Aronen HJ, Karonen JO, Liu Y, Nuutinen J, Wald LL, Sorensen AG
J Magn Reson Imaging 27(4): 866-71 (2008) [pdf]
Analytical methods of atherosclerosis research
Wang HH, Wang XF
Current Development in Atherosclerosis Research, 33-66, Nova Science Publishing, NY (2006).
Modeling atherosclerosis
Wang HH, Wang XF
Trends in Atherosclerosis Research, 279-311, Nova Science Publishing, NY, (2004).
Analytical models of atherosclerosis
Wang HH
Atherosclerosis 159: 1-7 (2001) [pdf]